Last week the results of the procurement exercise for 2015-2018 NHS national core content information resources was revealed.One of the pieces of inexplicable news in this announcement was that CINAHL was going to be discontinued unless the 2nd round of procurement decisions gave it a reprieve ( – fingers crossed, and a great overview by Alan Fricker covers all the detail). The second bombshell (from my point of view) was that Medline via OvidSP was going to stop, and the NHS were going to move to Proquest as a provider of Medline. Now, perhaps on the face of it this isn’t a big deal – most NHS ATHENS users will access their databases via the HDAS interface (Healthcare Databases Advanced Search) from the www.evidence.nhs.uk site. The fundamental reason that this interface was built was so that there’d be continuity for all users if/when content providers changed.
As a librarian who develops systematic review search strategies quite regularly the loss of Medline via OvidSP is catastrophic. This interface allows for sophisticated search strategies. (By this I mean strategies that often stretch well over 50 lines, and which contain multiple lines with adj* combinations. They contain a mixture of MeSH and freetext terms, and being able to export 400 papers at a time to Endnote makes life a lot easier when there are 20,000+ hits.) OvidSP is widely considered the search interface of choice for systematic reviews – eg The Cochrane Handbook offer search filters tailored for Medline via OvidSP in particular as well as the freely available PubMed.
Added to this is the fact that every experience I have ever had of using the Proquest interface has been deeply frustrating. I have to search ASSIA via this interface for any systematic reviews that require it. The interface is ok in principle, but the search algorithms can’t cope with anything terribly complicated and the export option crashes with extraordinary frequency (I have been in touch with Proquest helpdesk but with no improvement of functionality).
There are plenty of papers that have compared different interfaces, most recently:
I’m in the lucky position of currently having access to Medline via at least 4 interfaces (actually I’ve got access to medline via at least 8 interfaces, but lets not complicate things too much!):
- OvidSP using NHS ATHENS login
- HDAS using NHS ATHENS login
- Proquest using University of Cambridge login
- Pubmed (freely available)
 This gives me a great opportunity to have a taste of what I and any other NHS librarian and NHS ATHENS user is going to get in May 2015 onwards (assuming that NHS ATHENS users don’t have to wait for ages while the techies at NICE try to make Medline in HDAS interface with Proquest nicely….. )
This gives me a great opportunity to have a taste of what I and any other NHS librarian and NHS ATHENS user is going to get in May 2015 onwards (assuming that NHS ATHENS users don’t have to wait for ages while the techies at NICE try to make Medline in HDAS interface with Proquest nicely….. )
I thought I’d look at the interfaces for Medline with the following points of comparison
- the number of hits for “type in the words and press go” searches
- the number of hits for TITLE only freetext searches
- the number of hits for MeSH only searches
- the speed at which it would export results to Endnote
so……
“type in the words and press go” searches
I started off with “type in the words and press go” searches – I used diabetes as my test word. Really, just typed in diabetes and pressed search.
Each database reacts to that in a different way – I knew that already.
- HDAS assumes you want to find the work in Title and/or Abstract.
- OvidSP works much harder: .mp. means the database will look in the folllowing fields: title, abstract, original title, name of substance word, subject heading word, keyword heading word, protocol supplementary concept word, rare disease supplementary concept word, unique identifier – clearly a much broader search
- Pubmed assumes you want: “diabetes mellitus”[MeSH Terms] OR (“diabetes”[All Fields] AND “mellitus”[All Fields]) OR “diabetes mellitus”[All Fields] OR “diabetes”[All Fields] OR “diabetes insipidus”[MeSH Terms] OR (“diabetes”[All Fields] AND “insipidus”[All Fields]) OR “diabetes insipidus”[All Fields]
- Proquest assumes.. I actually have no idea – but considering that the first paper on my list contained the word diabetes in the address of the 2nd author, I’m going to assume it’s searching in “all fields”
So, the number of hits for each was:
- HDAS – 350614
- OvidSP – 47354
- Pubmed – 487126
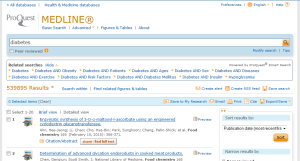
- Proquest – 539895
- my goodness – what additional content is Proqest searching to get so many more hits than the other interfaces? Is it a good thing that it’s being so much more sensitive than any of the other interfaces?
TITLE only freetext searches
- HDAS – 159145
- OvidSP – 159415
- Pubmed – 155728
- Proquest – 163263
The fact that HDAS and OvidSP get the same number of hits is exactly as I would expect – OvidSP is the source of HDAS content. The fact that Pubmed is more than either of them isn’t surprising either – (MGill University Healthcare Libraries have written a lovely overview of the differences between medline via Pubmed and medline via OvidSP) The fact that Proquest found significantly more again was interesting.
MeSH only searches
I exploded the MeSH terms.
- HDAS – 327890
- OvidSP – 327890
- Pubmed – 314182
- Proquest – 314038
So again, a difference. HDAS and OvidSP – both equal, and both gave the highest number of hits. Pubmed gave fewer, and Proquest the least – eh? what’s going on? why the reversal of sensitivity for Proquest?
What I didn’t take the time to establish was whether the Proquest hits were a pure subset of the Pubmed results, whether Pubmed results were a pure subset of the OvidSP/HDAS hits.
export results to Endnote
I test ran an export of 200 results from each database.
I took a screencast video to show the results in real time – it’s not an interesting video to watch unless you’re a *properly* nerdy librarian, but OvidSP and Pubmed were both winners – super quick – ie they took less than 20 seconds to export into Endnote. OvidSP gets the prize though because I’ve been able to change the settings to allow me to export 400 references at a time (any NHS ATHENS administrator can do this) – so effectively using OvidSP as my interface will always take half the time.
HDAS took c.50 seconds to export 200 references (after the 20 seconds I had to wait for the search results to display). One minute per 200 papers is dull, but bearable-ish depending on the number of hits.
However, after the first 1000 papers you have to slow down. From now on, you have to display each page of hits, select all the results, move to the next page, and repeat for up to 200 references then you can export this new set of 200. And repeat repeatedly depending on how many hits you get. Now things start to get significantly less bearable.
Proquest was marginally better than HDAS in that it allows for 100 articles to be displayed per page, but was the least reliable of the 4 interfaces in that it crashed twice when trying to direct export to endnote, and only succeeded when using the RIS export option. So great if I can get the references out, but that is by no means guaranteed.
Conclusions and areas for further consideration
So this was a very rough comparison, only looking at numbers of hits. It highlights that there are differences between the results of each interface (we knew that already) but it serves as a reminder why it’s so important to know the interface when the methods of a systematic review are written up.
The difference in the number of hits per search does worry me – but I can’t decide whether more hits is a better result than fewer – is lots of extra false positives better than the potential of missing relevant papers? Hopefully your search strategy would be sufficiently sensitive in itself to be able to balance out any peculiarity of the interface (ie you’d never just search for one word, or just use MeSH in a systematic review – you’d use a combination of multiple MeSH and multiple synonymous freetext terms).
Second, I think the speed of the export to endnote from OvidSP is very impressive – and makes a dull task in any systematic review significantly less time consuming. So anything slower, less reliable, or that exports smaller numbers of references is bad news.
But what else should I have considered?
- results when combining a complex strategy – how well does each database cope with a strategy of c.50 lines?
- ability to use more than just and and or as search operators. Any systematic review (and many less complicated search strategies) need more sensitive proximity searches (eg cancer adj3 breast)
- perhaps most importantly for NHS ATHENS users (but impossible for me at present to test) is the ability of HDAS to interface with Proquest, so that from May 2015 I’ll be able to get the same number of hits in the native interface as I would using HDAS.



Hi Isla, Thank you for this comparison! Really helpful. I just want to ask if there is a missing number in the Ovid´s results in “type in the words and press go” searches. The number is so much lower for Ovid. Greetings Marie